A new study led by a team of bioscientists from Durham University, UK, in collaboration with University of Liverpool, Northumbria University and New England Biolabs, hopes to exploit newly characterized defense systems in bacteria to compare changes to the human genome.
Undergraduates at Durham University have also been working on this research to demonstrate the complex workings of bacterial innate immunity.
Bacteria have evolved a multitude of defense systems to protect themselves from viruses called bacteriophages. Many of these systems have already been developed into useful biotechnological tools, such as for gene editing, where small changes are made to the target DNA.
The researchers demonstrated that two defense systems worked in a complementary manner to protect the bacteria from bacteriophages.
One system protected the bacteria from bacteriophages that did not have any modifications to their DNA.
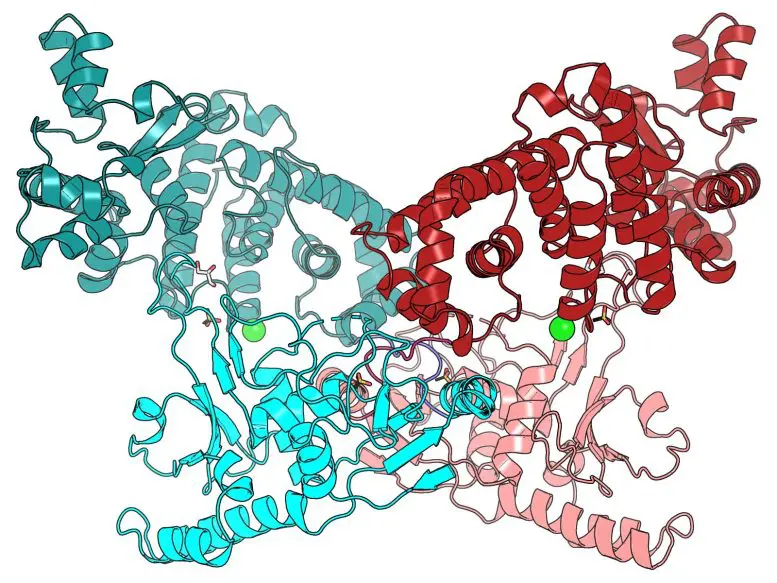
Some bacteriophages modify their DNA to avoid this first defense system. A second system, called BrxU, protected the bacteria from those bacteriophages with modified DNA, thereby providing a second layer of defense.
The researchers built an extremely detailed 3-D picture of BrxU to better understand how it protects from bacteriophages with modified DNA.
BrxU has the potential to be another useful biotechnological tool, because the same DNA modifications that BrxU recognizes appear throughout the human genome, and alter in cancer and neurodegenerative diseases.
Senior author of the study, Dr. Tim Blower, an Associate Professor and Lister Institute Prize Fellow in Durham University’s Department of Biosciences, said: “Being able to recognize modified DNA is crucial, as similar modifications are found throughout the DNA of the human genome.
“This extra layer of information, the “epigenome,” alters as we grow, and also changes in cases of cancer and neurodegenerative diseases.
“If we can develop BrxU as a biotechnological tool for mapping this epigenome, it will transform our understanding of the adaptive information controlling our growth and disease progression.”
The study findings from lead author Dr. David Picton and co-workers are published in the journal Nucleic Acids Research.
The 97 undergraduates involved in this work were in the final years of their BSc or MBiol degrees in the Department of Biosciences, Durham University.
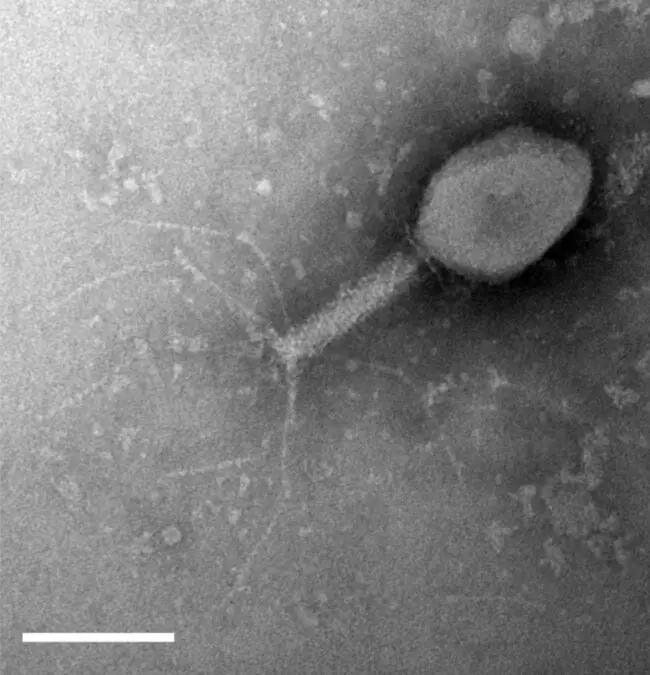
As part of a Microbiology Workshop designed to provide research-led teaching, they were tasked with isolating new bacteriophages for study. These bacteriophages thankfully don’t harm humans, but just as the human immune system responds to infections, bacteria have been forced to evolve their own immune systems that protect from bacteriophages.
Bacteriophages were collected from the River Wear, College ponds, and other waterways around Durham. They were then used to test the bacteriophage innate immunity in E. coli bacteria.
Reference: “The phage defence island of a multidrug resistant plasmid uses both BREX and type IV restriction for complementary protection from viruses” by David Picton, Yvette Luyten, Richard Morgan, Andrew Nelson, Darren Smith, David Dryden, Jay Hinton and Tim Blower, 17 October 2021, Nucleic Acids Research.
DOI: 10.1093/nar/gkab906
The research was funded in the UK by the Biotechnology and Biological Sciences Research Council Newcastle-Liverpool-Durham Doctoral Training Partnership, the Lister Institute of Preventive Medicine, Durham University’s Biophysical Sciences Institute, and the Wellcome Trust.